Abstract
Background: To satisfy rising biotechnological needs, climate change, and depleting water supplies, amylases that can survive high temperatures, and salt concentrations must be developed. Amylases are the most important enzymes that comprise approximately 25–33% of the international enzyme market and have a great role in many industries.
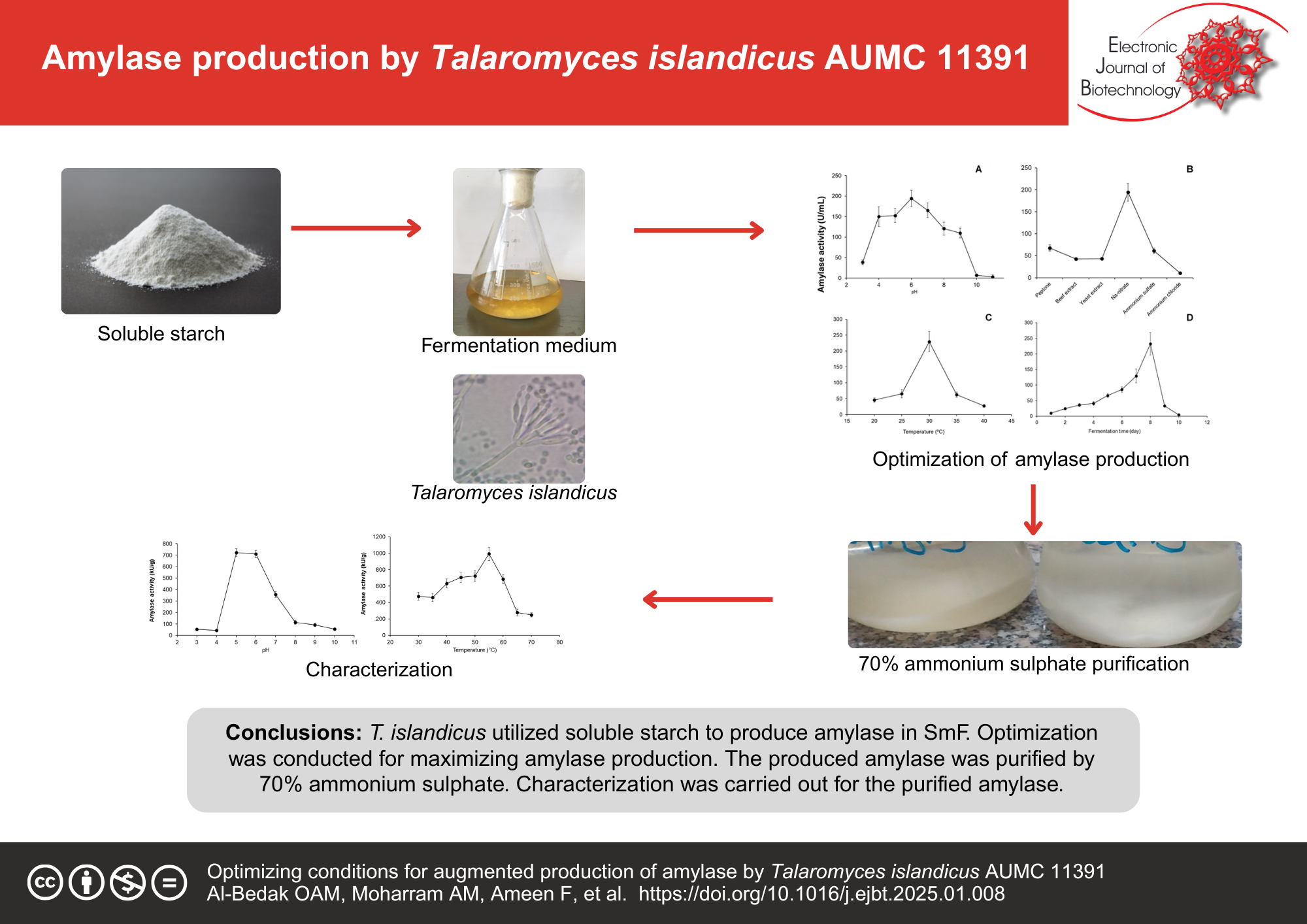
Results: In this investigation, Talaromyces islandicus AUMC 11391 was used to produce amylase in submerged fermentation (SmF) in an augmented concentration of 232 ± 36 U/mL after 8 d of incubation at pH 6.0 and 30°C using sodium nitrate as a nitrogen supply. The obtained enzyme was partly purified employing 70% ammonium sulfate and then dialysis. The activity of the produced amylase exhibited reached the peak (992.14 ± 80 U/mg protein) at pH 5.0 and 55°C. Cu, Co, Fe, Ni, Ca, and Zn had an activating effect on the activity of the amylase enzyme at pH 5.0 and 55°C by 134.57, 123.1, 115.4, 109.76, 105.43, and 103.2%, respectively. The Km and Vmax were recorded as 132.1 mM and 60.6 µmol/min, respectively. T. islandicus AUMC 11391′s-amylase hydrolyzed the raw starch of maize, sorghum, wheat, rice, and oat at rates of 12.3, 113.7, 32.25, 34.67, and 73.6% in contrast to the soluble starch.
Conclusions: This enhanced α-amylase from T. islandicus AUMC 11391 looks to be a potential option to meet the present amylase requirements of a variety of industrial processes because of its improved production and beneficial properties.
References
Arora N, Kaur S, Kaur S. Use of agro industrial residue for the production of amylase by Penicillium sp. for application in food industry. J Biotechnol Biomaterials 2017;7(2):256. https://doi.org/10.4172/2155-952X.1000256
Abd El-Latif H, Heba G, Abd El-Fatah BE, et al. Molecular identification of naturally isolated Candida tropicalis AUN-H100 and optimization of their extracellular amylase production. Int J Agr Sci Vet Med 2019;7(4):28-34.
Al-Bedak OA, Sakr RS, AL-Kolaibe AM. The microbial amylases: an overview with practical consequences and applications. J. Mcrobial Exp. 2022;10(4):130-4.
Abdel-Mageed HM, Fouad SA, Teaima MH, et al. Optimization of nano spray drying parameters for production of ?-amylase nanopowder for biotheraputic applications using factorial design. Drying Technology 2019;37(16):2152-60. https://doi.org/10.1080/07373937.2019.1565576
Saini R, Saini HS, Dahiya A. Amylases: Characteristics and industrial applications. J Pharmacog Phytochem 2017;6(4):1865-71.
Farooq MA, Ali S, Hassan A, et al. Biosynthesis and industrial applications of ?-amylase: A review. Archev Microbiol 2021;203(4):1281-92. https://doi.org/10.1007/s00203-020-02128-y PMid: 33481073
Pereira ADS, Fontan RIDC, Franco M, et al. Study of alpha-amylase obtained by solid state fermentation of cassava residue in aqueous two-phase systems. Braz J Chem Eng 2018;35(3):1141-52. https://doi.org/10.1590/0104-6632.20180353s20170003
Shruthi BR, Achur RNH, Boramuthi TN. Optimized solid-state fermentation medium enhances the multienzymes production from Penicillium citrinum and Aspergillus clavatus. Curr Microbiol 2020;77(9):2192-206. https://doi.org/10.1007/s00284-020-02036-w PMid: 32451686
Kuddus M. Enzymes in food biotechnology: Production, Applications and Future Prospects. Academic Press; 2019. https://doi.org/10.1016/C2016-0-04555-2
Maghraby YR, El-Shabasy RM, Ibrahim AH, et al. Enzyme immobilization technologies and industrial applications. ACS Omega 2023;8(6):5184-96. https://doi.org/10.1021/acsomega.2c07560 PMid: 36816672
Golden DA, Beuchat LR, Brackett RE. Direct plating technique for enumeration of Listeria monocytogenes in foods. J Assoc Offic Anal Chem 1988;71(3):647-50. https://doi.org/10.1093/jaoac/71.3.647
Smith D, Onions AH. The preservation and maintenance of living fungi, CAB international; 1994, 132 p. https://doi.org/10.1079/9780851989020.0000
Moharram AM, Yasser MM, Sayed M, et al. Mycobiota and mycotoxins contaminating rice grains in El-Minia, Governorate, Egypt. Biosci Biotechnol Res Asia 2019;16(1):167-78. https://doi.org/10.13005/bbra/2734
Moubasher AH, Ismail MA, Al-Bedak OA, et al. Ramophialophora chlamydospora, a new species from an alkaline lake of Wadi-El-Natron, Egypt. Asian J Mycol 2019;2(1):110-7. https://doi.org/10.5943/ajom/2/1/5
Al-Bedak OA, Moubasher AH. Aspergillus gaarensis, a new addition to section Circumdati from soil of Lake El-Gaar in Wadi-El-Natron, Egypt. Stud Fung 2020;5(1):59-65. https://doi.org/10.5943/sif/5/1/5
Innis MA, Gelfand DH, Sninsky JJ, et al. PCR protocols: A guide to methods and applications. Academic Press Inc. 1989.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molec Biol Evol 2013;30(4):772-80. https://doi.org/10.1093/molbev/mst010 PMid: 23329690
Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol 2010;10:210. https://doi.org/10.1186/1471-2148-10-210 PMid: 20626897
Kumar S, Stecher G, Li M, et al. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molec Biol Evol 2018;35(6):1547. https://doi.org/10.1093/molbev/msy096 PMid: 29722887
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evol 1985;39(4):783-791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x PMid: 28561359
Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinf 1998;14(9):817-18. https://doi.org/10.1093/bioinformatics/14.9.817 PMid: 9918953
Al-Bedak OA, Teama E, Ali E, et al. Impact of fumigation with phosphine on viability of wheat grains stored for six months at two levels of moisture content, in addition to description of four new records of associated fungi and assessment of their potential for enzymatic production. J Basic Appl Mycol (Egypt) 2020;11:77-97.
Al-Bedak OA, Moharram AM, Hussein NAH, et al. Microbial exploitation of feather wastes for sustainable production of keratinase and collagenase enzymes by Didymella keratinophila AUMC 15399 in submerged fermentation. Ferment 2023;9(6):507. https://doi.org/10.3390/fermentation9060507
Ramadan AM, Shehata RM, El-Sheikh HH, et al. Exploitation of sugarcane bagasse and environmentally sustainable production, purification, characterization, and application of lovastatin by Aspergillus terreus AUMC 15760 under solid-state conditions. Molecul 2023;28(10):4048. https://doi.org/10.3390/molecules28104048 PMid: 37241788
Moharram AM, Zohri A-NA, Hesham AE-L, et al. Production of cold-active pectinases by three novel Cladosporium species isolated from Egypt and application of the most active enzyme. Sci Rep 2022;12(1):15599. https://doi.org/10.1038/s41598-022-19807-z PMid: 36114347
Dangkulwanich M, Kongnithigarn K, Aurnoppakhun N. Colorimetric measurements of amylase activity: Improved accuracy and efficiency with a smartphone. J Chem Ed 2018;95(1):141-5. https://doi.org/10.1021/acs.jchemed.7b00468
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 1959;31(3):426-8. https://doi.org/10.1021/ac60147a030
Lowry OH, Rosebrough NJ, Lewis Farr A,et al. Measurement with the folin phenol reagent. J Biol Chem. 1951;193(1):265-75. https://doi.org/10.1016/S0021-9258(19)52451-6 PMid: 14907713
Lineweaver H, Burk D. The determination of enzyme dissociation constants. J Am Chem Soc 1934;56(3):658-66. https://doi.org/10.1021/ja01318a036
Moubasher AH, Ismail MA, Mohamed RA, et al. Production and purification of extreme xylanase from Aspergillus flavus AUMC 10331 in sub-merged fermentation. Eur J Biol Res 2019;9(1):20-8. http://dx.doi.org/10.5281/zenodo.2586103
Ismail MA, Moubasher AH, Mohamed RA, et al. Agro-industrial residues as alternative sources for cellulases and xylanases production and purification of xylanase produced by Aspergillus flavus AUMC 10331 isolated from extreme habitat. Curr Res Environ Appl Mycol 2018;8(3):313-22. https://doi.org/10.5943/cream/8/3/3
Al-Kolaibe AMG, Moharram AM, Al-Bedak OA. Worthwhile enzyme production and eco-friendly bioconversion of three agricultural residues by Aspergillus curvatus and Aspergillus gaarensis, promising enzyme-producers isolated from extreme environment. J Basic Appl Mycol (Egypt) 2021;12:1-14.
Moharram AM, Zohri A, Hesham A, et al. Production of cocktail enzymes by three Cladosporium isolates and bioconversion of orange peel wastes into valuable enzymes. J Pure Appl. Microbiol 2021;15(4):2336-46. https://doi.org/10.22207/JPAM.15.4.58
Alwakeel SS, Ameen F, Al Gwaiz H, et al. Keratinases produced by Aspergillus stelliformis, Aspergillus sydowii, and Fusarium brachygibbosum isolated from human hair: yield and activity. J Fung 2021;7(6):471. https://doi.org/10.3390/jof7060471 PMid: 34200943
Jujjavarapu SE, Dhagat S. Evolutionary trends in industrial production of ?-amylase. Recent Pat Biotechnol 2019;13(1):4-18. https://doi.org/10.2174/2211550107666180816093436 PMid: 30810102
Pandey A, Soccol CR, Nigam P, et al. Biotechnological potential of agro-industrial residues. I: sugarcane bagasse. Biores Technol 2000;74(1):69-80. https://doi.org/10.1016/S0960-8524(99)00142-X
Ashok PP, Dasgupta D, Ray A, et al. Challenges and prospects of microbial ?-amylases for industrial application: A review. World J Microbiol Biotechnol 2024;40(2):44. https://doi.org/10.1007/s11274-023-03821-y PMid: 38114825
Sahu PK, Singh R, Shrivastava M, et al. Microbial production of ?-amylase from agro-waste: An approach towards biorefinery and bio-economy. Energy Nexus 2024;14:100293. https://doi.org/10.1016/j.nexus.2024.100293
Mohamed SA, Al-Malki AL, Kumosani TA. Partial purification and characterization of five ?-amylases from a wheat local variety (Balady) during germination. Austral J Basic Appl Sci 2009;3(3):1740-8.
Nouadri T, Meraihi Z, Shahrazed DD, et al. Purification and characterization of the ?-amylase isolated from Penicillium camemberti PL21. Afr. J. Biochem. Res 2010;4(6):155-62.
Sethi BK, Dikshit B, Sahoo SL, et al. Extracellular production of amylase and protease by Penicillium purpurogenum BKS9. Biosci Bioeng 2017;5(1):1-6. https://doi.org/10.13189/bb.2017.050101
Ekedegba FE, Ogbonna AI, NwibariBMW, et al. Optimization of amylase production in three fungal species. Asian J Biochem Gen Mol Biol 2022;12(4):1-9. https://doi.org/10.9734/ajbgmb/2022/v12i4266
Sethi S, Gupta S. Isolation, characterization and optimization of cultural conditions for amylase production from fungi. J Global Sci 2015;4(9):3356-63.
Jain D, Katyal P. Optimization of gluco-amylase production from Aspergillus sp. for its use in saccharification of liquefied corn starch. 3 Biotech 2018;8:101. https://doi.org/10.1007/s13205-018-1131-4 PMid: 29430363
Ogbonna CN, Nnaji OB, Chioke OJ. Isolation of amylase and cellulase producing fungi from decaying tubers and optimization of their enzyme production in solid and submerged cultures. Int J Biotechnol Food Sci 2018;6(1):9-17.
Zhao F, Li Y, Li C, et al. Exo-type, endo-type and debranching amylolytic enzymes regulate breadmaking and storage qualities of gluten-free bread. Carb Poly 2022;298:120124. https://doi.org/10.1016/j.carbpol.2022.120124 PMid: 36241296
Adeoyo O, Adewumi O. Characterization of ?-amylase and antimicrobial activity of Penicillium chrysogenum, J BioSci Biotechnol 2024;13(1):23-8.
Jakheng S, Thomas J, Umar M, et al. Screening of locally isolated Penicillium species from the soil for amylase production. J Appl Life Sci Int 2020;23(12):76-83. https://doi.org/10.9734/jalsi/2020/v23i1230207
Hon KJY, Amran NAAM, Abdullah AN. Comparative analysis of ?-amylase sequences in selected Aspergillus species and analysis of amylase activity in three locally isolated Aspergillus species. Sci Technol Asia 2024;29(3):224-243.
Anguraj A, Michael HSR, Sivaraman RK, et al. Bioprospecting of fungi to produce protease and amylase. In: Uppuluri KB, Selvasembian R, editors, Bioprospecting of multi-tasking fungi for a sustainable environment. Springer, Singapore. 2024:169-98. https://doi.org/10.1007/978-981-97-4113-7_8
Asghar S, Pervez S, Bibi A, et al. Optimization of production conditions for ?-amylase as an important biodesizing agent. Int Conf Biol Res Appl Sci 2024;79-80.
Al-Agamy MH, Alhuzani MR, Kelany MS, et al. Production and partial characterization of ?-amylase enzyme from marine actinomycetes. BioMed Res Int 2021;2021(1):5289848. https://doi.org/10.1155/2021/5289848 PMid: 34917683
Pol D, Laxman RS, Rao M. Purification and biochemical characterization of endoglucanase from Penicillium pinophilum MS 20. Indian J Biochem Biophys 2012;49(3):189-94. PMid: 22803334
Maharana AK. Extracellular cold active endoglucanase and pigment producing psychrotolerant Penicillium pinophilum. Int J Pharm Pharmaceut Sci 2016;8(10):164-9. https://doi.org/10.22159/ijpps.2016v8i10.13441
de Castro AM, de Carvalho ML, Leite SGF, et al. Cellulases from Penicillium funiculosum: Production, properties and application to cellulose hydrolysis, J Ind Microbiol Biotechnol 2010;37(2):151-8. https://doi.org/10.1007/s10295-009-0656-2 PMid: 19902281
Balkan B, Ertan F. Production and properties of ??amylase from Penicillium chrysogenum and its application in starch hydrolysis. Prep Biochem Biotechnol 2005;35(2):169-78. https://doi.org/10.1081/PB-200054740 PMid: 15881598
Sindhu R, Suprabha G, Shashidhar S. Optimization of process parameters for the production of ?-amylase from Penicillium janthinellum (NCIM 4960) under solid state fermentation. Adv J Microbiol Res 2009;3(9):498-503.
Erdal S, Taskin M. Production of ?-amylase by Penicillium expansum MT-1 in solid-state fermentation using waste Loquat (Eriobotrya japonica Lindley) kernels as substrate. Roman Biotechnol Lett 2010;15(3):5342-50.
Lopes PHS, Pasin TM, Benassi VM, et al. Standardization of the cultivation of Rhizopus arrhizus using agroindustrial residues: High production of amylases in pineapple peel. Braz Arch Biol Technol 2024;67:e24240293. https://doi.org/10.1590/1678-4324-2024240293
Castro D, Marques ASC, Almeida MR, et al. L-asparaginase production review: Bioprocess design and biochemical characteristics. Appl Microbiol Biotechnol 2021;105:4515-34. https://doi.org/10.1007/s00253-021-11359-y PMid: 34059941
Doyle EM, Kelly CT, Fogarty WM. The high maltose-producing ?-amylase of Penicillium expansum. Appl Microbiol Biotechnol 1989;30:492-6. https://doi.org/10.1007/BF00263854
Saha BC, Silman RW, Bothast RJ. Amylolytic enzymes produced by a color variant strain of Aureobasidium pullulans. Curr Microbiol 1993;26:267-73. https://doi.org/10.1007/BF01575916
Abou-Zeid A. Production, purification and characterization of an extracellular alpha-amylase enzyme isolated from Aspergillus flavus. Microbios 1997;89(358):55-66. PMid: 9218355
Mohamed SA, Azhar EI, Ba-Akdah MM, et al. Production, purification and characterization of ?-amylase from Trichoderma harzianum grown on mandarin peel. Afr J Microbiol Res 2011;5(7):930-40. https://doi.org/10.5897/AJMR10.890
Viswanathan S, Rohini S, Rajesh R, et al. Production and medium optimization of amylase by Bacillus spp. using submerged fermentation method. World J Chem 2014;9(1):01-06.
Al-Johani NB, Al-Seeni MN, Ahmed YM. Optimization of alkaline ?-amylase production by thermophilic Bacillus subtilis. Afr J Trad Compl Alt Med 2017;14(1):288-301. https://doi.org/10.21010/ajtcam.v14i1.31 PMid: 28480407
Upgade A, Nandeshwar A, Samant L. Assessment of fungal protease enzyme from French bean using A. niger by Solid State Fermentation. J Microbiol Biotechnol Res 2011;1(4):45-51.
Bhargav S, Panda BP, Ali M, et al. Solid-state fermentation: An overview. Chem Biochem Eng Quart 2008;22(1):49-70.
Du R, Song Q, Zhang Q, et al. Purification and characterization of novel thermostable and Ca-independent ?-amylase produced by Bacillus amyloliquefaciens BH072. Int J Biol Macromol 2018;115:1151-6. https://doi.org/10.1016/j.ijbiomac.2018.05.004 PMid: 29729336
Raul D, Biswas T, Mukhopadhyay S, et al. Production and partial purification of alpha amylase from Bacillus subtilis (MTCC 121) using solid state fermentation. Biochem Res Int 2014;2014(1):568141. https://doi.org/10.1155/2014/568141 PMid: 24672727
Wu X, Wang Y, Tong B, et al. Purification and biochemical characterization of a thermostable and acid-stable alpha-amylase from Bacillus licheniformis B4-423. Int J Biol Macromol 2018;109:329-37. https://doi.org/10.1016/j.ijbiomac.2017.12.004 PMid: 29233713
Dash BK, Rahman MM, Sarker PK. Molecular identification of a newly isolated Bacillus subtilis BI19 and optimization of production conditions for enhanced production of extracellular amylase. BioMed Res Int 2015;2015(1):859805. https://doi.org/10.1155/2015/859805 PMid: 26180814
Dar RA, Saba I, Shahnawaz M, et al. Isolation, purification and characterization of carboxymethyl cellulase (CMCase) from endophytic Fusarium oxysporum producing podophyllotoxin. Adv Enz Res 2013;1(04):91-6. https://doi.org/10.4236/aer.2013.14010
Ali I, Akbar A, Anwar M, et al. Purification and characterization of a polyextremophilic ?-amylase from an obligate halophilic Aspergillus penicillioides isolate and its potential for souse with detergents. BioMed Res Int 2015;2015:245649. https://doi.org/10.1155/2015/245649 PMid: 26180787
Mohapatra B, Banerjee U, Bapuji M. Characterization of a fungal amylase from Mucor sp. associated with the marine sponge Spirastrella sp. J Biotechnol 1998;60(1-2):113-7. https://doi.org/10.1016/S0168-1656(97)00197-1
Aquino A, Jorge JA, Terenzi HF, et al. Studies on a thermostable ?-amylase from the thermophilic fungus Scytalidium thermophilum. Appl Microbiol Biotechnol 2003;61:323-8. https://doi.org/10.1007/s00253-003-1290-y PMid: 12743761
Varalakshmi K, Kumudini B, Nandini B, et al. Production and characterization of alpha-amylase from Aspergillus niger JGI 24 Isolated in Bangalore. Polish J Microbiol 2009;58(1):29-36. PMid: 19469283
Metin K, Koc O, Ate?lier B, et al. Purification and characterization of ?-amylase produced by Penicillium citrinum HBF62. Afr J Biotechnol 2010;9(45):7692-701.
Zaferanloo B, Bhattacharjee S, Ghorbani MM, et al. Amylase production by Preussia minima, a fungus of endophytic origin: Optimization of fermentation conditions and analysis of fungal secretome by LC-MS. BMC Microbiol 2014;14(1):55. https://doi.org/10.1186/1471-2180-14-55 PMid: 24602289
Xu QS, Yan YS, Feng JX. Efficient hydrolysis of raw starch and ethanol fermentation: A novel raw starch-digesting glucoamylase from Penicillium oxalicum. Biotechnol Biofuels 2016;9(1):216. https://doi.org/10.1186/s13068-016-0636-5 PMid: 27777618
Savic S, Savic S, Petrovic S, et al. Activity and stability of dextranase from new Penicillium funiculosum TFZ.91: Optimization by response surface methods. Iran J Sci Technol, Trans A: Sci 2022;46(3):747-760. https://doi.org/10.1007/s40995-022-01293-7
Zhao H. Effect of ions and other compatible solutes on enzyme activity, and its implication for biocatalysis using ionic liquids. J Mol Cat B: Enz 2005;37(1-6):16-25. https://doi.org/10.1016/j.molcatb.2005.08.007

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Copyright (c) 2025 Electronic Journal of Biotechnology