Abstract
Background: In cancer, the process of anoikis is intimately associated with the emergence and progression. N6-methyladenosine modification and m6A modification play an important role in regulating long non-coding RNAs. The liver hepatocellular carcinoma patients’ data, including clinical and prognostic data, were obtained via The Cancer Genome Atlas database. The univariate, multivariate Cox and Least Absolute Selection Operator (LASSO) regression were performed to gain anoikis- and m6A-related lncRNAs. The Kaplan-Meier method was employed to assess the overall survival rate for groups of high- and low risks.
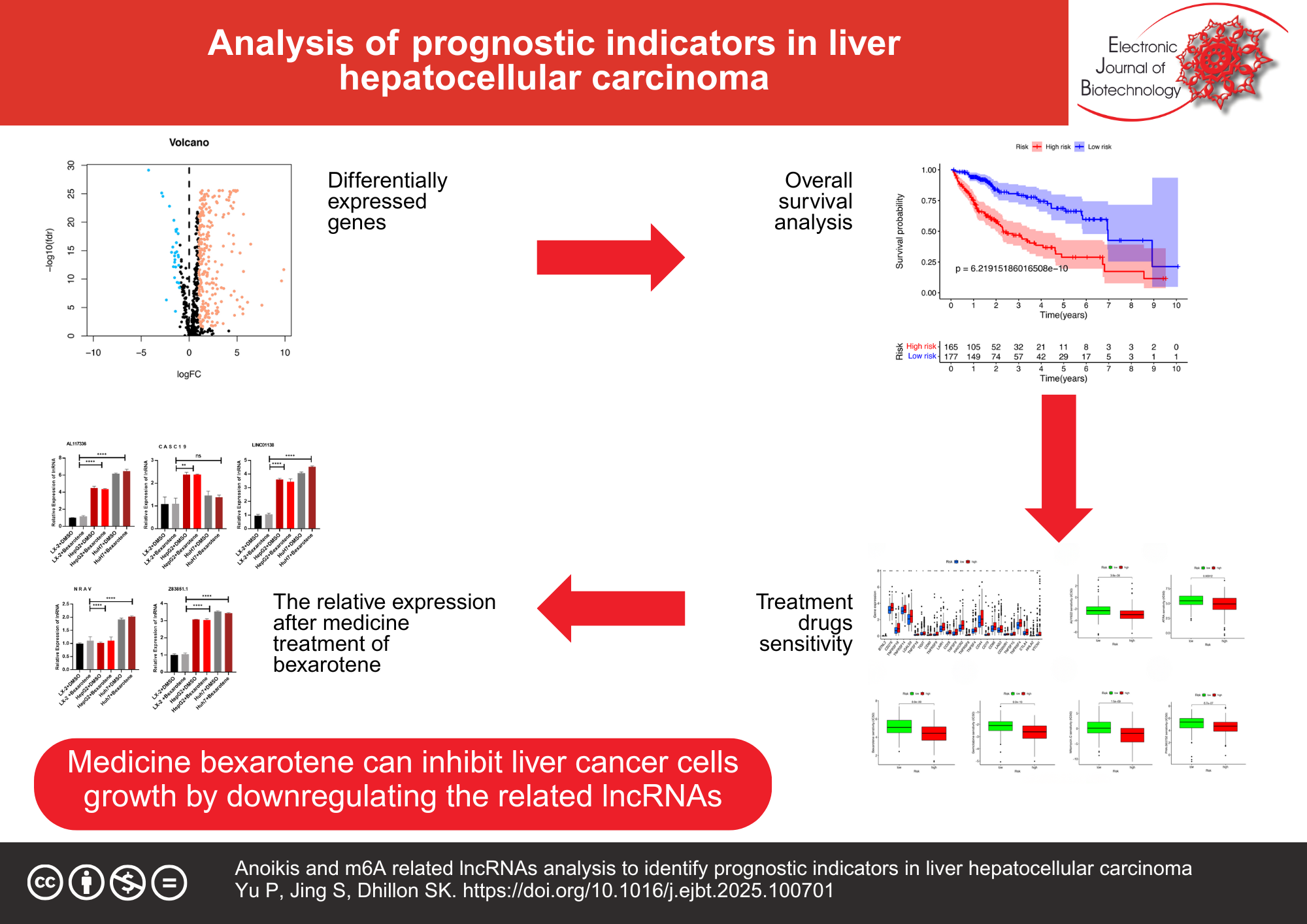
Results: A signature comprising six anoikis- and m6A-related lncRNAs was constructed: AL117336.3, LINC01138, Z83851.1, NRAV, CASC19 and AC009283.1. The clinicopathological variables, the anoikis- and m6A-related lncRNA signature demonstrated superior diagnostic efficacy, with an area under the receiver operating characteristic curve of 0.810. In the high-risk group, the overall survival was shown to be inferior to that of in group of low risk, while patients were classified by distinct clinicopathological variables. The ssGSEA and CIBERSORT immune analysis demonstrated that the predictive signature was significantly associated with liver cancer patients’ immune status. The chemotherapy drugs ATRA, AUY922, bexarotene, gemcitabine, mitomycin-C, and PHA have been found to have greater sensitivity in treating high-risk patients. qRT-PCR showed that Z83851.1, NRAV and CASC19 lncRNAs were associated with poor prognosis and were high-risk factors. AC009283.1 lncRNA may have anti-cancer properties.
Conclusions: The predictive signature is capable of independently predicting the prognosis of liver cancer patients for understanding the mechanisms of anoikis- and m6A-related lncRNAs in liver hepatocellular carcinoma and offering clinical guidance to patients with liver cancer.
References
Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893-917. https://doi.org/10.1002/ijc.25516 PMid: 21351269
Raza A, Sood GK. Hepatocellular carcinoma review: Current treatment, and evidence-based medicine. World J Gastroenterol. 2014;20(15):4115-27. https://doi.org/10.3748/wjg.v20.i15.4115
PMid: 24764650
Kishi Y, Hasegawa K, Sugawara Y, et al. Hepatocellular carcinoma: current management and future development-improved outcomes with surgical resection. Int J Hepatol. 2011;2011(1):728103. https://doi.org/10.4061/2011/728103 PMid: 21994868
Jiang Y, Zhao X, Fu J, et al. Progress and challenges in precise treatment of tumors with PD-1/PD-L1 blockade. Front Immunol. 2020;11:339. https://doi.org/10.3389/fimmu.2020.00339. PMid: 32226426
Haslam A, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open. 2019;2(5):e192535. https://doi.org/10.1001/jamanetworkopen.2019.2535 PMid: 31050774
Liu JW, Supandi F, Dhillon SK. Ferroptosis-related long noncoding RNA signature predicts prognosis of clear cell renal carcinoma. Folia Biol (Praha). 2022;68(1):1-15. https://doi.org/10.14712/fb2022068010001 PMid: 36201853
Taddei ML, Giannoni E, Fiaschi T, et al. Anoikis: An emerging hallmark in health and diseases. J Pathol. 2012;226(2):380-93. https://doi.org/10.1002/path.3000 PMid: 21953325
Yu Y, Liu B, Li X, et al. ATF4/CEMIP/PKC? promotes anoikis resistance by enhancing protective autophagy in prostate cancer cells. Cell Death Dis. 2022;13(1):46. https://doi.org/10.1038/s41419-021-04494-x PMid: 35013120
Shimokawa M, Yoshizumi T, Itoh S, et al. Modulation of Nqo1 activity intercepts anoikis resistance and reduces metastatic potential of hepatocellular carcinoma. Cancer Sci. 2020;111(4):1228-40. https://doi.org/10.1111/cas.14320 PMid: 31968140
Michel JB. Anoikis in the cardiovascular system: Known and unknown extracellular mediators. Arterioscler Thromb Vasc Biol. 2003;23(12):2146-54. https://doi.org/10.1161/01.ATV.0000099882.52647.E4 PMid: 14551156
Madajewski B, Boatman MA, Chakrabarti G, et al. Depleting tumor-NQO1 Potentiates anoikis and inhibits growth of NSCLC. Mol Cancer Res. 2016;14(1):14-25. https://doi.org/10.1158/1541-7786.MCR-15-0207-T PMid: 26553038
Du S, Miao J, Zhu Z, et al. NADPH oxidase 4 regulates anoikis resistance of gastric cancer cells through the generation of reactive oxygen species and the induction of EGFR. Cell Death Dis. 2018;9(10):948. https://doi.org/10.1038/s41419-018-0953-7 PMid: 30237423
Han J, Yu J, Dai Y, et al. Overexpression of miR-361-5p in triple-negative breast cancer (TNBC) inhibits migration and invasion by targeting RQCD1 and inhibiting the EGFR/PI3K/Akt pathway. Biomol Biomed. 2019;19(1):52-9. https://doi.org/10.17305/bjbms.2018.3399 PMid: 29924958
Jiang X, Liu B, Nie Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):74. https://doi.org/10.1038/s41392-020-00450-x PMid: 33611339
Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15(5):293-306. https://doi.org/10.1038/nrg3724 PMid: 24662220
Lan T, Li H, Zhang D, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18(1):186. https://doi.org/10.1186/s12943-019-1106-z PMid: 31856849
Chen Y, Peng C, Chen J, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18(1):127. https://doi.org/10.1186/s12943-019-1053-8 PMid: 31438961
Wei C, Wang B, Peng D, et al. Pan-cancer analysis shows that ALKBH5 is a potential prognostic and immunotherapeutic biomarker for multiple cancer types including gliomas. Front Immunol. 2022;13:849592. https://doi.org/10.3389/fimmu.2022.849592 PMid: 35444654
Wang H, Liang L, Dong Q, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-?B pathway in hepatocellular carcinoma. Theranostics. 2018;8(10):2814-29. https://doi.org/10.7150/thno.23012 PMid: 29774077
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452-63. https://doi.org/10.1016/j.ccell.2016.03.010 PMid: 27070700
Chi Y, Wang D, Wang J, et al. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):1015. https://doi.org/10.3390/cells8091015 PMid: 31480503
Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661-7. https://doi.org/10.1038/onc.2017.184 PMid: 28604750
Hou C, Wu X, Li C, et al. A cuproptosis-associated long non-coding RNA signature for the prognosis and immunotherapy of lung squamous cell carcinoma. Biomol Biomed. 2023;23(4):624-33. https://doi.org/10.17305/bb.2022.8481 PMid: 36724022
Cai HJ, Zhuang ZC, Wu Y, et al. Development and validation of a ferroptosis-related lncRNAs prognosis signature in colon cancer. Biomol Biomed. 2021;21(5):569-76. https://doi.org/10.17305/bjbms.2020.5617
Stelzer G, Dalah I, Stein TI, et al. In-silico human genomics with GeneCards. Human Genomics. 2011;5(6):709. https://doi.org/10.1186/1479-7364-5-6-709 PMid: 22155609
Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-50. https://doi.org/10.1073/pnas.0506580102 PMid: 16199517
Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48-61. http://doi.org/10.1016/j.cell.2014.12.033 PMid: 25594174
Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453-7. https://doi.org/10.1038/nmeth.3337 PMid: 25822800
Hakimi AA, Reznik E, Lee CH, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29(1):104-16. https://doi.org/10.1016/j.ccell.2015.12.004 PMid: 26766592
Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35-44. https://doi.org/10.1016/j.cell.2016.02.065 PMid: 26997480
Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207-11. https://doi.org/10.1126/science.aad0095 PMid: 26359337
Maeser D, Gruener RF, Huang RS. oncoPredict: An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22(6): bbab260 . https://doi.org/10.1093/bib/bbab260 PMid: 34260682
Geeleher P, Cox N, Huang RS. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. https://doi.org/10.1371/journal.pone.0107468 PMid: 25229481
Anwanwan D, Singh SK, Singh S, et al. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873(1):188314. https://doi.org/10.1016/j.bbcan.2019.188314 PMid: 31682895
Amini M, Looha MA, Zarean E, et al. Global pattern of trends in incidence, mortality, and mortality-to-incidence ratio rates related to liver cancer, 1990-2019: A longitudinal analysis based on the global burden of disease study. BMC Public Health. 2022;22(1):604. https://doi.org/10.1186/s12889-022-12867-w PMid: 35351047
Galicia-Moreno M, Silva-Gomez JA, Lucano-Landeros S, et al. Liver cancer: Therapeutic challenges and the importance of experimental models. Can J Gastroenterol Hepatol. 2021;202181):8837811. https://doi.org/10.1155/2021/8837811 PMid: 33728291
Tan K, Goldstein D, Crowe P, et al. Uncovering a key to the process of metastasis in human cancers: a review of critical regulators of anoikis. J Cancer Res Clin Oncol. 2013;139(11):1795-805. https://doi.org/10.1007/s00432-013-1482-5 PMid: 23912151
Chiarugi P, Giannoni E. Anoikis: a necessary death program for anchorage-dependent cells. Biochem Pharmacol. 2008;76(11):1352-64. https://doi.org/10.1016/j.bcp.2008.07.023 PMid: 18708031
Cao Z, Livas T, Kyprianou N. Anoikis and EMT: Lethal "Liaisons" during cancer progression. Crit Rev Oncog. 2016;21(3-4):155-68. https://doi.org/10.1615/CritRevOncog.2016016955 PMid: 27915969
Wang J, Luo Z, Lin L, et al. Anoikis-associated lung cancer metastasis: Mechanisms and therapies. Cancers. 2022;14(19):4791. https://doi.org/10.3390/cancers14194791 PMid: 36230714
Du S, Cao K, Wang Z, et al. Comprehensive analysis of anoikis-related lncRNAs for predicting prognosis and response of immunotherapy in hepatocellular carcinoma. IET Syst Biol. 2023;17(4):198-211. https://doi.org/10.1049/syb2.12070 PMid: 37417684
Wang WH, Hullinger RL, Andrisani OM. Hepatitis B virus X protein via the p38MAPK pathway induces E2F1 release and ATR kinase activation mediating p53 apoptosis. J Biol Chem. 2008;283(37):25455-67. https://doi.org/10.1074/jbc.M801934200 PMid: 18606816
Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov. 2014;4(4):405-14. https://doi.org/10.1158/2159-8290.CD-13-0136 PMid: 24658082
Mattick JS, Amaral PP, Carninci P, et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24(6):430-47. https://doi.org/10.1038/s41580-022-00566-8 PMid: 36596869
Jiang X, Gao YL, Li JY, et al. An anoikis-related lncRNA signature is a useful tool for predicting the prognosis of patients with lung adenocarcinoma. Heliyon. 2023;9(11):e22200. https://doi.org/10.1016/j.heliyon.2023.e22200 PMid: 38053861
Meng WJ, Guo JM, Huang L, et al. Anoikis-related long non-coding RNA signatures to predict prognosis and immune infiltration of gastric cancer. Bioengineering. 2024;11(9):893. https://doi.org/10.3390/bioengineering11090893 PMid: 39329635
Lu Q, Wang L, Gao Y, et al. lncRNA APOC1P1-3 promoting anoikis-resistance of breast cancer cells. Cancer Cell Int. 2021;21(1):232. https://doi.org/10.1186/s12935-021-01916-w PMid: 33902604
Qiao FH, Tu M, Liu HY. Role of MALAT1 in gynecological cancers: Pathologic and therapeutic aspects. Oncol Lett. 2021;21(4):333. https://doi.org/10.3892/ol.2021.12594 PMid: 33692865
Zhang J, Li Q, Xue B, et al. MALAT1 inhibits the Wnt/?-catenin signaling pathway in colon cancer cells and affects cell proliferation and apoptosis. Biomolecules and Biomedicine. 2020;20(3):357-64. https://doi.org/10.17305/bjbms.2019.4408
Qu CX, Shi XC, Zai LQ, et al. LncRNA CASC19 promotes the proliferation, migration and invasion of non-small cell lung carcinoma via regulating miRNA-130b-3p. Eur Rev Med Pharmacol Sci. 2019;23(3 Suppl):247-55. https://doi.org/10.26355/eurrev_201908_18654 PMid: 31389608
Wang JJ, Li XM, He L, et al. Expression and function of long non-coding RNA CASC19 in colorectal cancer. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2017;39(6):756-61. https://doi.org/10.3881/j.issn.1000-503x.2017.06.004 PMid: 29338818
Wang WJ, Guo CA, Li R, et al. Long non-coding RNA CASC19 is associated with the progression and prognosis of advanced gastric cancer. Aging. 2019;11(15):5829-47. http://doi.org/10.18632/aging.102190
Wang XD, Lu J, Lin YS, et al. Functional role of long non-coding RNA CASC19/miR-140-5p/CEMIP axis in colorectal cancer progression in vitro. World J Gastroenterol. 2019;25(14):1697-714. https://doi.org/10.3748/wjg.v25.i14.1697 PMid: 31011255
Lv J, Fan HX, Zhao XP, et al. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016;382(2):166-75. https://doi.org/10.1016/j.canlet.2016.08.029 PMid: 27597739
Hou Y, Tang Y, Ma C, et al. Overexpression of CASC19 contributes to tumor progression and predicts poor prognosis after radical resection in hepatocellular carcinoma. Dig Liver Dis. 2023;55(6):799-806. https://doi.org/10.1016/j.dld.2022.12.001 PMid: 36805849
Cedro-Tanda A, Ríos-Romero M, Romero-Córdoba S, et al. A lncRNA landscape in breast cancer reveals a potential role for AC009283.1 in proliferation and apoptosis in HER2-enriched subtype. Sci Rep. 2020;10(1):13146. https://doi.org/10.1038/s41598-020-69905-z PMid: 32753692
Li Z, Zhang J, Liu X, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. 2018;9(1):1572. https://doi.org/10.1038/s41467-018-04006-0 PMid: 29679004
You M, Xie Z, Zhang N, et al. Signaling pathways in cancer metabolism: Mechanisms and therapeutic targets. Signal Transduct Target Ther. 2023;8(1):196. https://doi.org/10.1038/s41392-023-01442-3 PMid: 37164974
Yin J, Ren W, Huang X, et al. Potential Mechanisms connecting purine metabolism and cancer therapy. Front Immunol. 2018;9:1697. https://doi.org/10.3389/fimmu.2018.01697 PMid: 30105018
Siddiqui A, Ceppi P. A non-proliferative role of pyrimidine metabolism in cancer. Mol Metab. 2020;35:100962. https://doi.org/10.1016/j.molmet.2020.02.005 PMid: 32244187
Goodall GJ, Wickramasinghe VO. RNA in cancer. Nature Reviews Cancer. 2021;21(1):22-36. https://doi.org/10.1038/s41568-020-00306-0 PMid: 33082563
Liang R, Cheng A, Lu S, et al. Seleno-amino acid metabolism reshapes the tumor microenvironment: from cytotoxicity to immunotherapy. Int J Biol Sci. 2024;20(7):2779-89. https://doi.org/10.7150/ijbs.95484 PMid: 38725849
Giles JR, Globig AM, Kaech SM, et al. CD8+ Tcells in the cancer-immunity cycle. Immunity. 2023;56(10):2231-53. https://doi.org/10.1016/j.immuni.2023.09.005 PMid: 37820583
Cazzetta V, Franzese S, Carenza C, et al. Natural killer-dendritic cell interactions in liver cancer: Implications for immunotherapy. Cancers. 2021;13(9):2184. https://doi.org/10.3390/cancers13092184 PMid: 34062821
Arvanitakis K, Mitroulis I, Germanidis G. Tumor-associated neutrophils in hepatocellular carcinoma pathogenesis, prognosis, and therapy. Cancers. 2021;13(12):2899. https://doi.org/10.3390/cancers13122899 PMid: 34200529
Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat Rev Immunol. 2020;20(11):651-68. https://doi.org/10.1038/s41577-020-0306-5 PMid: 32433532
Sakharkar MK, Shashni B, Sharma K, et al. Therapeutic implications of targeting energy metabolism in breast cancer. PPAR Res. 2013;2013(1):109285. https://doi.org/10.1155/2013/109285 PMid: 23431283

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Copyright (c) 2026 Electronic Journal of Biotechnology