Abstract
Background: The ability of Bacillus safensis to synthesize xylanase and other industrially important enzymes utilizing lignocellulosic biomass makes it advantageous for a variety of biotechnology applications. Thus, the current investigation aimed to optimize conditions and medium components for maximizing xylanase production by a newly isolated Bacillus safensis strain using banana rachis (peel of banana tree) as a novel source of carbon.
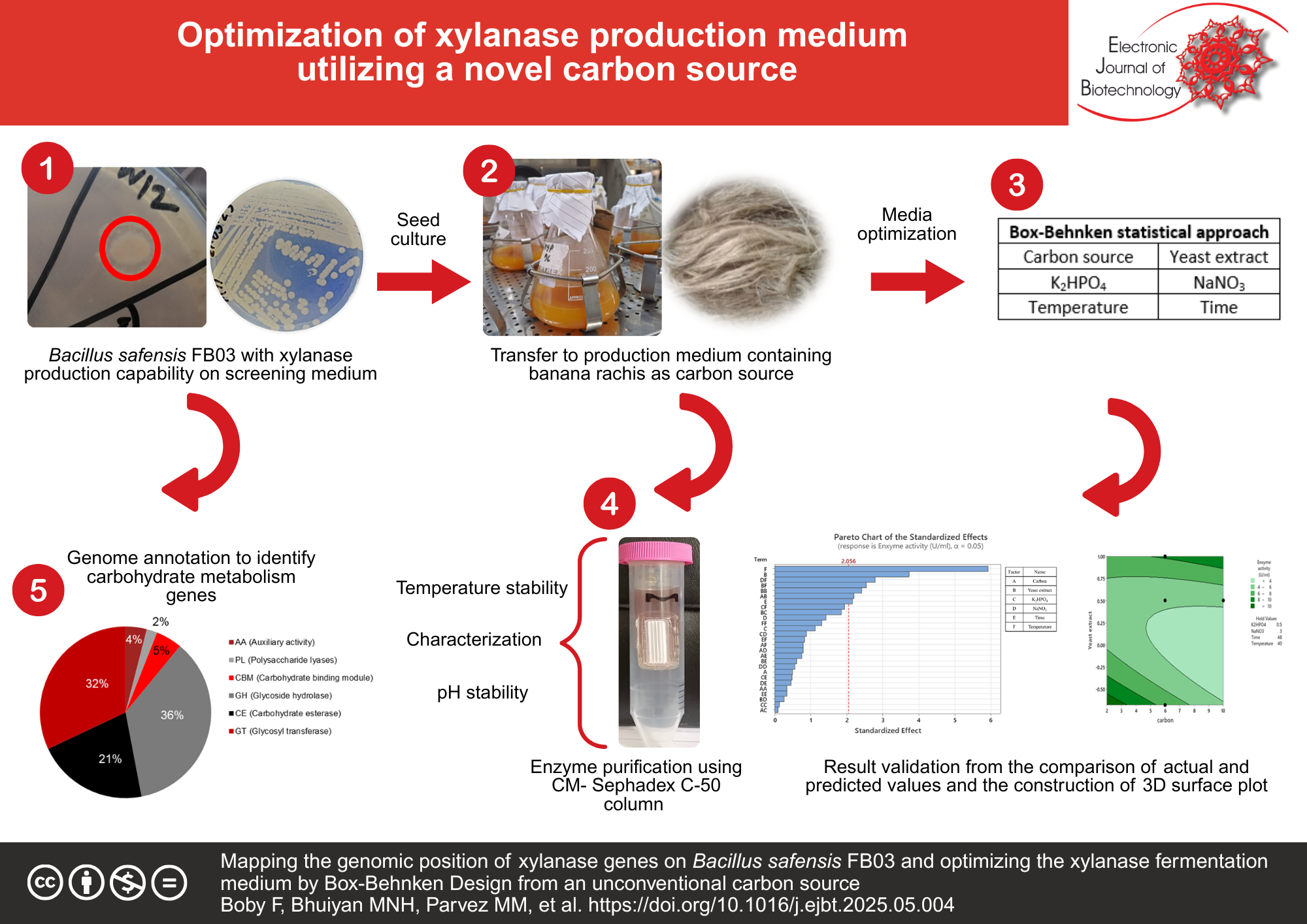
Results: Upon employing Box-Behnken Design (BBD) statistical approach, the highest enzyme activity was obtained 25.24 U/ml at 2 g/L banana rachis, 1 g/L yeast extract, 1 g/L K2HPO4, 5 g/L NaNO3, 35°C and 72 h of incubation time. The purified enzyme showed 10 times higher enzyme activity (143.6 U/ml) with 2.3 mg/ml protein concentration. The enzyme was found to maintain stability up to 60°C in a wide range of pH (6 to 10). Analysis of whole genome sequencing data revealed the presence of xylanase production and xylan metabolic genes (xynA, xynB, xylP, xylT) on Bacillus safensis FB03. Also, from genome annotation, different carbohydrate metabolic genes such as glycoside hydrolases (GHs), glycosyl transferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), auxiliary activities (AAs), and carbohydrate binding modules (CBMs) were identified.
Conclusions: In accordance with our research, banana rachis can be considered as a major medium component to develop an economical fermentation process for the production of xylanase by Bacillus safensis FB03. Additionally, identification of the genomic location of xyl genes provides valuable insight towards genetic engineering for the development of a more potent industrial strain.
References
Rastogi, M., Shrivastava, S., Shukla, P. Bioprospecting of xylanase producing fungal strains: Multilocus phylogenetic analysis and enzyme activity profiling. Journal of Basic Microbiology, 2022;62(2):150–161. https://doi.org/10.1002/jobm.202100408 PMid: 34783043
Chaudhary, R., Pandey, A., Tiwari, R., et al. Current status of xylanase for biofuel production: A review on classification and characterization. Biomass Conversion and Biorefinery, 2021;13(10):8773–8791. https://doi.org/10.1007/s13399-021-01948-2
Walia, A., Guleria, S., Mehta, P., et al. Microbial xylanases and their industrial application in pulp and paper biobleaching: A review. 3 Biotech, 2017;7(1):11. https://doi.org/10.1007/s13205-016-0584-6 PMid: 28391477
Shrivastava, S. Introduction to glycoside hydrolases: Classification, identification and occurrence. In: Industrial Applications of Glycoside Hydrolases. Springer, Singapore 2020:3–84. https://doi.org/10.1007/978-981-15-4767-6_1
Dias, L. M., dos Santos, B. V., Albuquerque, C. J. B., et al. Biomass sorghum as a novel substrate in solid-state fermentation for the production of hemicellulases and cellulases by Aspergillus niger and A. fumigatus. Journal of Applied Microbiology, 2018;124(3:708–718. https://doi.org/10.1111/jam.13672 PMid: 29253315
Bhardwaj, N., Kumar, B., Verma, P. A detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresources and Bioprocessing, 2019;6(1):40. https://doi.org/10.1186/s40643-019-0276-2
Parab, P., Khandeparker, R. Xylanolytic enzyme consortia from Bacillus sp. NIORKP76 for improved biobleaching of kraft pulp. Bioprocess and Biosystems Engineering, 2021;44(12):2513–2524. https://doi.org/10.1007/s00449-021-02623-6 PMid: 34402971
Contesini, F.J., de Melo, R.R., Sato, H.H. An overview of Bacillus proteases: From production to application. Critical Reviews in Biotechnology, 2018:38(3):321–334. https://doi.org/10.1080/07388551.2017.1354354 PMid: 28789570
Rastogi, M., Shrivastava, S. Recent advances in second generation bioethanol production: An insight to pretreatment, saccharification and fermentation processes. Renewable and Sustainable Energy Reviews, 2017;80:330–340. https://doi.org/10.1016/j.rser.2017.05.225
Lateef, A., Adelere, I.A., Gueguim-Kana, E.B. The biology and potential biotechnological applications of Bacillus safensis. Biologia, 2015;70(4):411–419. https://doi.org/10.1515/biolog-2015-0062
Devi, S., Dwivedi, D., Bhatt, A.K. Utilization of agroresidues for the production of xylanase by Bacillus safensis XPS7 and optimization of production parameters. Fermentation, 2022;8(5):221. https://doi.org/10.3390/fermentation8050221
Rekik, H., Jaouadi N.Z., Gardouri F, et al. Production, purification and biochemical characterization of a novel detergent-stable serine alkaline protease from Bacillus safensis strain RH12. International Journal of Biological Macromolecules, 2019;121:1227–1239. https://doi.org/10.1016/j.ijbiomac.2018.10.139 PMid: 30352229
Patel, K., Parikh, S. Identification, production, and purification of a novel lipase from Bacillus safensis. Journal of Applied Biology and Biotechnology, 2022;10(4):73–76. https://doi.org/10.7324/JABB.2022.100410
Pandya, U., Saraf, M. Purification and characterization of antifungal chitinase from Bacillus safensis MBCU6 and its application for production of chito-oligosaccharides. Biologia 2015;70(7):863–868. https://doi.org/10.1515/biolog-2015-0112
Sharma, H., Singh, J., Batra, N. Isolation and molecular identification of ?-amylase producing bacteria from hot water spring by 16S rRNA gene sequencing. Indian Journal of Experimental Biology, 2019;57:937-944.
Singh, R.S., Singh, R.P. Response surface optimization of endoinulinase production from a cost effective substrate by Bacillus safensis AS-08 for hydrolysis of inulin. Biocatalysis and Agricultural Biotechnology, 2014;3(4):365–372. https://doi.org/10.1016/j.bcab.2014.05.002
Nath, A., Sarkar, S., Maitra, M., et al. An experimental study on production of intracellular ?-galactosidase at different conditions by batch process using isolated Bacillus safensis (JUCHE 1) and characterization of synthesized ?-galactosidase. Journal of the Institution of Engineers (India): Series E, 2012;93(2):55–60. https://doi.org/10.1007/s40034-013-0011-z
Al-Shawi, A.H. Enhanced xylanase production from Bacillus safensis MABS6 using sorghum straw substrate: Optimization, characterization, and biotechnological applications. Catrina: The International Journal of Environmental Sciences, 2024;29(1):9–24. https://doi.org/10.21608/cat.2023.239574.1209
Lateef, A., AdelereI A., Gueguim-Kana, E.B. Bacillus safensis LAU 13: A new source of keratinase and its multi-functional biocatalytic applications. Biotechnology & Biotechnological Equipment, 2015;29(1):54–63. https://doi.org/10.1080/13102818.2014.986360 PMid: 26740788
Azeem, M.A., Shah FH, Ullah A, et al. Biochemical characterization of halotolerant Bacillus safensis PM22 and its potential to enhance growth of maize under salinity stress. Plants, 2022;11(13):1721. https://doi.org/10.3390/plants11131721 PMid: 35807673
Zagorchev, L., Kamenova, P., Odjakova, M. The role of plant cell wall proteins in response to salt stress. The Scientific World Journal, 2014;2014(1):764089. https://doi.org/10.1155/2014/764089 PMid: 24574917
Wu, P.S., Liu, C.H., Hu, S.Y. Probiotic Bacillus safensis NPUST1 administration improves growth performance, gut microbiota, and innate immunity against Streptococcus iniae in Nile tilapia (Oreochromis niloticus). Microorganisms, 2021;9(12):2494. https://doi.org/10.3390/microorganisms9122494 PMid: 34946096
Kwon, E.H., Adhikari A, Imran M, et al. Novel melatonin-producing Bacillus safensis EH143 mitigates salt and cadmium stress in soybean. Journal of Pineal Research, 2024;76(4):e12957. https://doi.org/10.1111/jpi.12957 PMid: 38803089
Priyalaxmi, R., Murugan, A., Raja, P., et al. Bioremediation of cadmium by Bacillus safensis (JX126862), a marine bacterium isolated from mangrove sediments. International Journal of Current Microbiology and Applied Sciences, 2014;3(12):326-335.
Kamble, R.D., Jadhav, A.R. Isolation, purification, and characterization of xylanase produced by a new species of Bacillus in solid state fermentation. International Journal of Microbiology, 2012;2012(1):683193. https://doi.org/10.1155/2012/683193 PMid: 22315613
Samanta, A.K., Kolte, A.P., Senani, S., et al. A simple and efficient diffusion technique for assay of endo ?-1,4-xylanase activity. Brazilian Journal of Microbiology, 2011;42(4):1349–1353. https://doi.org/10.1590/S1517-83822011000400016 PMid: 24031763
Bhalla, A., Bischoff, K.M., Sani, R.K. Highly thermostable xylanase production from a thermophilic Geobacillus sp. strain WSUCF1 utilizing lignocellulosic biomass. Frontiers in Bioengineering and Biotechnology, 2015;3:84. https://doi.org/10.3389/fbioe.2015.00084 PMid: 26137456
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 1976;72(1–2):248–254. https://doi.org/10.1016/0003-2697(76)90527-3 PMid: 942051
Grant, J.R., Enns E, Matinier E, et at. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Research, 2023;51(W1):W484–W492. https://doi.org/10.1093/nar/gkad326 PMid: 37140037
Drula, E., Garron, M. L., Dogan, S., et al. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Research,2022;50(D1):D571–D577. https://doi.org/10.1093/nar/gkab1045 PMid: 34850161
Lodi, R.S., Dong X., Jiang C, et al. Antimicrobial activity and enzymatic analysis of endophytes isolated from Codonopsis pilosula. FEMS Microbiology Ecology, 2023;99(8):fiad071. https://doi.org/10.1093/femsec/fiad071 PMid: 37365694
Tarafdar, A., Sirohi R, Gaur VK, et al. Engineering interventions in enzyme production: Lab to industrial scale. Bioresource Technology, 2021;326:124771. https://doi.org/10.1016/j.biortech.2021.124771 PMid: 33550211
Marimuthu, M., Sorimuthu, A., Muruganantham, S. Production and optimization of xylanase enzyme from Bacillus subtilis using agricultural wastes by solid state fermentation. International Journal of Pharmaceutical Investigation, 2019;9(4):169–173.
Al Mousa, A.A., Abo-Dahab, N.F., Hassane, A.M.A., et al. Harnessing Mucor spp. for xylanase production: Statistical optimization in submerged fermentation using agro-industrial wastes. BioMed Research International, 2022;2022(1):3816010. https://doi.org/10.1155/2022/3816010 PMid: 35496057
Abdella, A., Segato, F., Wilkins, M.R. Optimization of process parameters and fermentation strategy for xylanase production in a stirred tank reactor using a mutant Aspergillus nidulans strain. Biotechnology Reports, 2020;26:e00457. https://doi.org/10.1016/j.btre.2020.e00457 PMid: 32420050
Güler, F., Özçelik, F. Screening of xylanase producing Bacillus species and optimization of xylanase process parameters in submerged fermentation. Biocatalysis and Agricultural Biotechnology, 2023;51:102801. https://doi.org/10.1016/j.bcab.2023.102801
Prabha, B., Ramesh, D., Sriramajayam, S., et al. Optimization of pyrolysis process parameters for fuel oil production from the thermal recycling of waste polypropylene grocery bags using the Box–Behnken design. Recycling, 2024;9(1):15. https://doi.org/10.3390/recycling9010015
Bakry, M.M., Salem, S.S., Atta, H.M., et al. Xylanase from thermotolerant Bacillus haynesii strain: Synthesis, characterization, optimization using Box-Behnken Design, and biobleaching activity. Biomass Conversion and Biorefinery, 2024;14(8):9779–9792. https://doi.org/10.1007/s13399-022-03043-6
Salmanizadeh, H., Beheshti-Maal, K., Nayeri, H., et al. Optimization of xylanase production by Pichia kudriavzevii and Candida tropicalis isolated from the wood product workshop. Brazilian Journal of Microbiology, 2024;55(1):155–168. https://doi.org/10.1007/s42770-023-01171-3 PMid: 37957443
Yadav, P., Maharjan J, Korpole S, et al. Production, purification, and characterization of thermostable alkaline xylanase from Anoxybacillus kamchatkensis NASTPD13. Frontiers in Bioengineering and Biotechnology, 2018;6:65. https://doi.org/10.3389/fbioe.2018.00065 PMid: 29868578
Sanghi, A., Garg, N., Gupta, V.K., et al. One-step purification and characterization of cellulase-free xylanase produced by alkalophilic Bacillus subtilis ash. Brazilian Journal of Microbiology, 2010;41(2):467-476. https://doi.org/10.1590/S1517-83822010000200029 PMid: 24031518
Ketsakhon, P., Thammasittirong, A., Thammasittirong, S.N.R. Adding value to rice straw waste for high-level xylanase production using a new isolate of Bacillus altitudinis RS3025. Folia Microbiologica, 2023;68(1):87–99. https://doi.org/10.1007/s12223-022-00998-x PMid: 35945409
Saleem, A., Waris, S., Ahmed, T., et al. Biochemical characterization and molecular docking of cloned xylanase gene from Bacillus subtilis RTS expressed in E. coli. International Journal of Biological Macromolecules, 2021;168:310–321. https://doi.org/10.1016/j.ijbiomac.2020.12.001 PMid: 33309670
Dhaver, P., Pletschke, B., Sithole, B., et al. Optimization, purification, and characterization of xylanase production by a newly isolated Trichoderma harzianum strain by a two-step statistical experimental design strategy. Scientific Reports, 2022;12(1):17791. https://doi.org/10.1038/s41598-022-22723-x PMid: 36273028
Gupta, G.K., Dixit, M., Kapoor, R K., et al. Xylanolytic enzymes in pulp and paper industry: New technologies and perspectives. Molecular Biotechnology, 2022;64(2):130–143. https://doi.org/10.1007/s12033-021-00396-7 PMid: 34580813
Nagar, S., Gupta, V.K., Kumar, D., et al. Production and optimization of cellulase-free, alkali-stable xylanase by Bacillus pumilus SV-85S in submerged fermentation. Journal of Industrial Microbiology and Biotechnology, 2010;37(1):71–83. https://doi.org/10.1007/s10295-009-0650-8 PMid: 19859753
Sepahy, A.A., Ghazi, S., Sepahy, M.A. Cost-effective production and optimization of alkaline xylanase by indigenous Bacillus mojavensis AG137 fermented on agricultural waste. Enzyme Research, 2011;2011(1):593624. PMid: 21904670
de Carvalho, L.M., Borelli G, Camargo AP, et al. Bioinformatics applied to biotechnology: A review towards bioenergy research. Biomass and Bioenergy, 2019;123:195–224. https://doi.org/10.1016/j.biombioe.2019.02.016
Deshmukh, R.A., Jagtap, S., Mandal, M.K., et al. Purification, biochemical characterization and structural modelling of alkali-stable ?-1,4-xylan xylanohydrolase from Aspergillus fumigatus R1 isolated from soil. BMC Biotechnology, 2016;16(1):11. https://doi.org/10.1186/s12896-016-0242-4 PMid: 26847222
Sharma, P.K., Chand, D. Purification and characterization of thermostable cellulase-free xylanase from Pseudomonas sp. XPB-6. Advances in Microbiology, 2012;2(1):17–25. https://doi.org/10.4236/aim.2012.21003
Halmschlag, B., Hoffmann K, Hanke R, et al. Comparison of isomerase and Weimberg pathway for ?-PGA production from xylose by engineered Bacillus subtilis. Frontiers in Bioengineering and Biotechnology, 2020;7:476. https://doi.org/10.3389/fbioe.2019.00476 PMid: 32039180
Mazmanian, S.K., Round, J.L., Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature, 2008;453(7195):620–625. https://doi.org/10.1038/nature07008 PMid: 18509436

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Copyright (c) 2025 Electronic Journal of Biotechnology