Abstract
Background: Biliary colic (BC), characterized by intermittent pain due to gallstone-related bile duct obstruction, remains poorly understood at the molecular level. Circulating exosomal microRNAs (miRNAs) have emerged as potential biomarkers for various diseases. This study aimed to identify exosomal miRNA profiles in BC patients and explore their therapeutic implications.
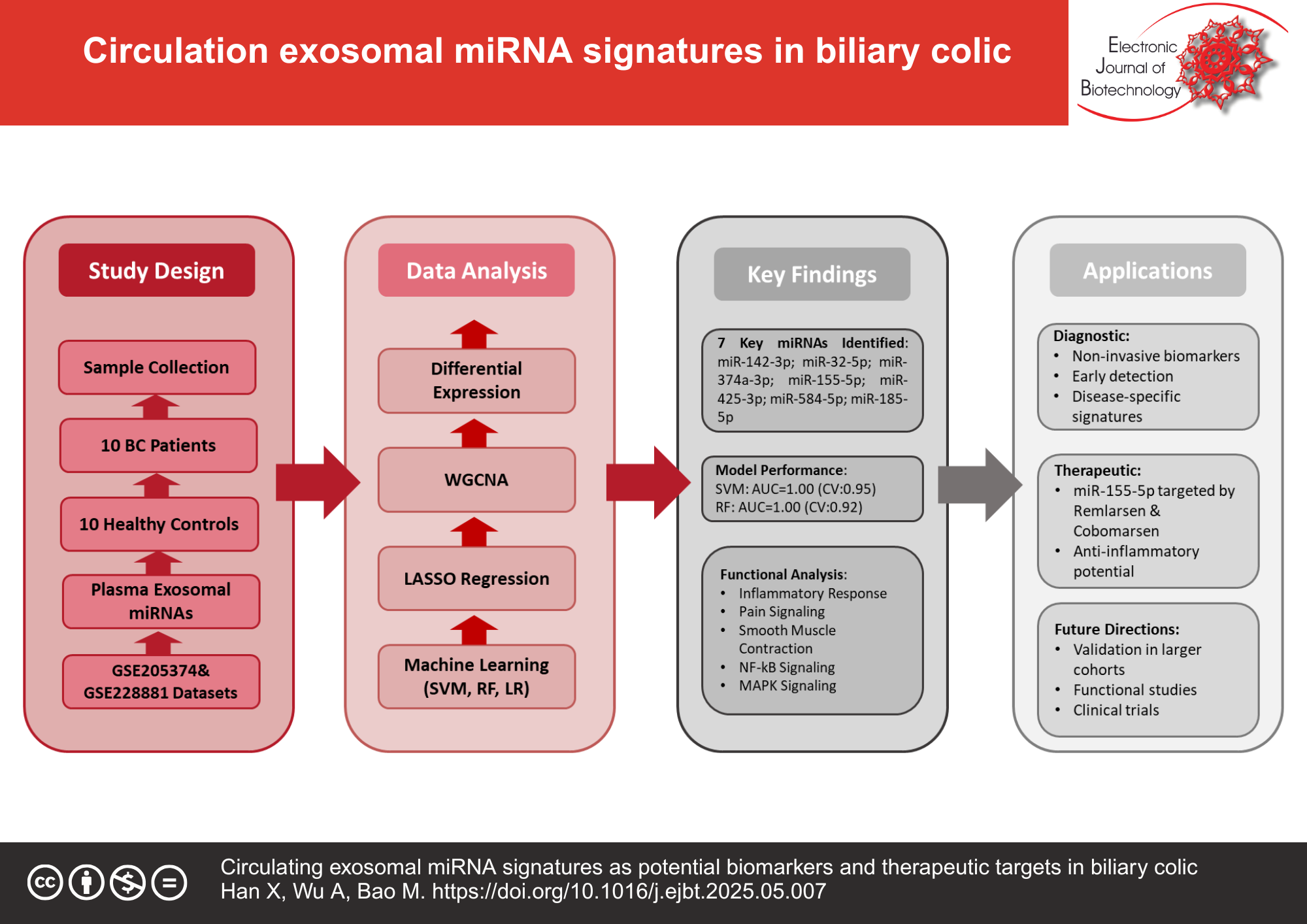
Results: Analysis of plasma exosomal miRNAs from 10 BC patients during acute attacks and 10 healthy controls (HCs) revealed distinct expression patterns separating BC from HC groups. Integration of differential expression analysis, WGCNA, and LASSO regression identified 7 key miRNAs (hsa-miR-142-3p, hsa-miR-32-5p, hsa-miR-374a-3p, hsa-miR-155-5p, hsa-miR-425-3p, hsa-miR-584-5p, hsa-miR-185-5p) strongly associated with BC. Support vector machine models using these miRNAs achieved excellent diagnostic performance (AUC = 1.0, where AUC represents Area Under the Curve). miRNA-targeting drugs including Remlarsen and Cobomarsen showed potential for therapeutic intervention.
Conclusions: This study identified specific exosomal miRNA signatures that distinguish BC patients from HC and revealed potential miRNA-targeting therapeutics. These findings advance our understanding of BC pathophysiology and provide direction for developing novel diagnostics and treatments.
References
Demehri FR, Alam HB. Evidence-based management of common gallstone-related emergencies. J Intensive Care Med 2014;31(1):3-13. https://doi.org/10.1177/0885066614554192 PMid: 25320159
Chen SY, Huang HY, Lin HP, et al. Piperlongumine induces autophagy in biliary cancer cells via reactive oxygen species-activated Erk signaling pathway. Int J Mol Med 2019;44(5):1687-1696. https://doi.org/10.3892/ijmm.2019.4324
Hucl T. Precursors to cholangiocarcinoma. Gastroenterol Res Pract 2019;2019(1):1389289. https://doi.org/10.1155/2019/1389289 PMid: 31814823
Guévremont D, Roy J, Cutfield NJ. et al. MicroRNAs in Parkinson’s disease: A systematic review and diagnostic accuracy meta-analysis. Sci Rep 2023;13:16272. https://doi.org/10.1038/s41598-023-43096-9
Huang S, Wang YJ, Guo J. Biofluid biomarkers of Alzheimer’s disease: progress, problems, and perspectives. Neurosci Bull 2022;38(6):677-691. https://doi.org/10.1007/s12264-022-00836-7 PMid: 35306613
Weidauer S, Wagner M, Hattingen E. White matter lesions in adults - a differential diagnostic approach. Rofo 2020;192(12):1154-1173. https://doi.org/10.1055/a-1207-1006 PMid: 32688424
Peterson MF, Otoc N, Sethi JK, et al. Integrated systems for exosome investigation. Methods 2015;87:31-45. https://doi.org/10.1016/j.ymeth.2015.04.015 PMid: 25916618
McBride JD, Rodriguez-Menocal L, Badiavas EV. Extracellular vesicles as biomarkers and therapeutics in dermatology: a focus on exosomes. J Invest Dermatol 2017;137(8):1622-1629. https://doi.org/10.1016/j.jid.2017.04.021 PMid: 28648952
Song X, Song Y, Zhang J, et al. Regulatory role of exosome-derived miRNAs and other contents in adipogenesis. Exp Cell Res 2024;441(1):114168. https://doi.org/10.1016/j.yexcr.2024.114168 PMid: 39004201
Kumar RMR. Exosomal microRNAs: impact on cancer detection, treatment, and monitoring. Clin Transl Oncol 2025;27(1):83-94. https://doi.org/10.1007/s12094-024-03590-6 PMid: 38971914
Ankasha SJ, Shafiee MN, Wahab NA, et al. Post-transcriptional regulation of microRNAs in cancer: From prediction to validation. Oncol Rev 2018;12(1):344. https://doi.org/10.4081/oncol.2018.344 PMid: 29989022
Tan PPS, Hall D, Chilian WM, et al. Exosomal microRNAs in the development of essential hypertension and its potential as biomarkers. Am J Physiol Heart Circ Physiol 2021;320(4):H1486-H1497. https://doi.org/10.1152/ajpheart.00888.2020 PMid: 33577433
Mishra PJ. MicroRNAs as promising biomarkers in cancer diagnostics. Biomark Res 2014;2:19. https://doi.org/10.1186/2050-7771-2-19 PMid: 25356314
Zafari S, Backes C, Petra Leidinger ,Meese E, et al. Regulatory microRNA networks: complex patterns of target pathways for disease-related and housekeeping microRNAs. Genomics Proteomics Bioinformatics 2015;13(3):159-68. https://doi.org/10.1016/j.gpb.2015.02.004 PMid: 26169798
Lin H, Shi X, Li H, et al. Urinary exosomal miRNAs as biomarkers of bladder cancer and experimental verification of mechanism of miR-93-5p in bladder cancer. BMC Cancer 2021;21(1):1293. https://doi.org/10.1186/s12885-021-08926-x PMid: 34861847
Yang P, Song F, Yang X, et al. Exosomal MicroRNA signature acts as an efficient biomarker for non-invasive diagnosis of gallbladder carcinoma. iScience 2022;25(9):104816. https://doi.org/10.1016/j.isci.2022.104816 PMid: 36043050
Arbelaiz A, Azkargorta M, Krawczyk M, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017;66(4):1125-1143. https://doi.org/10.1002/hep.29291 PMid: 28555885
Høgdall D, O'Rourke CJ, Larsen FO, et al. Whole blood microRNAs capture systemic reprogramming and have diagnostic potential in patients with biliary tract cancer. J Hepatol 2022;77(4):1047-1058. https://doi.org/10.1016/j.jhep.2022.05.036 PMid: 35750139
Taminau J, Meganck S, Lazar C, et al. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinformatics 2012;13:335. https://doi.org/10.1186/1471-2105-13-335 PMid: 23259851
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8(1):118-27. https://doi.org/10.1093/biostatistics/kxj037 PMid: 16632515
Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47. https://doi.org/10.1093/nar/gkv007 PMid: 25605792
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. https://doi.org/10.1186/1471-2105-9-559 PMid: 19114008
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32(18):2847-9. https://doi.org/10.1093/bioinformatics/btw313 PMid: 27207943
Cannon M, Stevenson J, Stahl K, et al. DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms. Nucleic Acids Res 2024;52(D1):D1227-D1235. https://doi.org/10.1093/nar/gkad1040 PMid: 37953380
Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020;367(6478):eaau6977. https://doi.org/10.1126/science.aau6977 PMid: 32029601
Li B, Cao Y, Sun M, et al. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J 2021;35(10):e21916. https://doi.org/10.1096/fj.202100294RR
Letelier P, Riquelme I, Hernández AH, et al. Circulating MicroRNAs as biomarkers in biliary tract cancers. Int J Mol Sci 2016;17(5):791. https://doi.org/10.3390/ijms17050791 PMid: 27223281
Supradit K, Wongprasert K, Tangphatsornruang S, et al. microRNA profiling of exosomes derived from plasma and their potential as biomarkers for Opisthorchis viverrini-associated cholangiocarcinoma. Acta Trop 2024;258:107362. https://doi.org/10.1016/j.actatropica.2024.107362 PMid: 39151716
Qin D, Wei R, Liu S, et al. A circulating miRNA-based scoring system established by WGCNA to predict colon cancer. Anal Cell Pathol 2019;2019(1):1571045. https://doi.org/10.1155/2019/1571045 PMid: 31871878
Guo L, Cai Y, Wang B, et al. Characterization of the circulating transcriptome expression profile and identification of novel miRNA biomarkers in hypertrophic cardiomyopathy. Eur J Med Res 2023;28(1):205. https://doi.org/10.1186/s40001-023-01159-7 PMid: 37391825
Zanoaga O, Braicu C, Chiroi P, et al. The role of miR-155 in nutrition: Modulating cancer-associated inflammation. Nutrients 2021;13(7):2245. https://doi.org/10.3390/nu13072245 PMid: 34210046
Mohan S, Hakami MA, Dailah HG, et al. From inflammation to metastasis: The central role of miR-155 in modulating NF-?B in cancer. Pathol Res Pract. 2024;253:154962. https://doi.org/10.1016/j.prp.2023.154962 PMid: 38006837
Abedi S, Behmanesh A, Mazhar FN, et al. Machine learning and experimental analyses identified miRNA expression models associated with metastatic osteosarcoma. Biochim Biophys Acta Mol Basis Dis. 2024;1870(7):167357. https://doi.org/10.1016/j.bbadis.2024.167357 PMid: 39033966
Aravind VA, Kouznetsova VL, Kesari S, et al. Using machine learning and miRNA for the diagnosis of esophageal cancer. J Appl Lab Med. 2024;9(4):684-95. https://doi.org/10.1093/jalm/jfae037 PMid: 38721901
He B, Zhao Z, Cai Q, et al. miRNA-based biomarkers, therapies, and resistance in cancer. Int J Biol Sci 2020;16(14):2628-2647. https://doi.org/10.7150/ijbs.47203 PMid: 32792861
Gallant-Behm CL, Piper J, Lynch JM, et al. A microRNA-29 mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J Invest Dermatol 2019;139(5):1073-81. https://doi.org/10.1016/j.jid.2018.11.007 PMid: 30472058
Anastasiadou E, Seto AG, Beatty X, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, slows DLBCL tumor cell growth in vitro and in vivo. Clin Cancer Res 2021;27(4):1139-1149. https://doi.org/10.1158/1078-0432.CCR-20-3139 PMid: 33208342
Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 2020;21(1):6. https://doi.org/10.1186/s12864-019-6413-7 PMid: 31898477
Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: Data mining, inference, and prediction. 2nd ed. New York: Springer; 2009. https://doi.org/10.1007/978-0-387-84858-7
Freitas AA. Investigating the role of Simpson's paradox in the analysis of top-ranked features in high-dimensional bioinformatics datasets. Brief Bioinform 2020;21(2):421-8. https://doi.org/10.1093/bib/bby126 PMid: 30629111
Ding Y, Tang J, Guo F. Identification of drug--target interactions via dual Laplacian regularized least squares with multiple kernel fusion. Knowl Based Syst 2020;204:106254. https://doi.org/10.1016/j.knosys.2020.106254
Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16(3):203-22. https://doi.org/10.1038/nrd.2016.246 PMid: 28209991
Wei L, Ding Y, Su R, et al. Prediction of human protein subcellular localization using deep learning. J Parallel Distrib Comput 2018;117:212-7. https://doi.org/10.1016/j.jpdc.2017.08.009

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Copyright (c) 2025 Electronic Journal of Biotechnology