Abstract
Background: Aquaculture has become the fastest-growing sector in recent decades, and this has led to intensified cultural practices to achieve high yields. However, such practices have raised concerns regarding their environmental impact, as aquaculture sludge and wastewater cause significant organic pollution. This study then aimed to isolate and screen proteolytic, amylolytic, and lipolytic bacteria and evaluate their consortia for enzyme production and bioremediation of shrimp pond sludge.
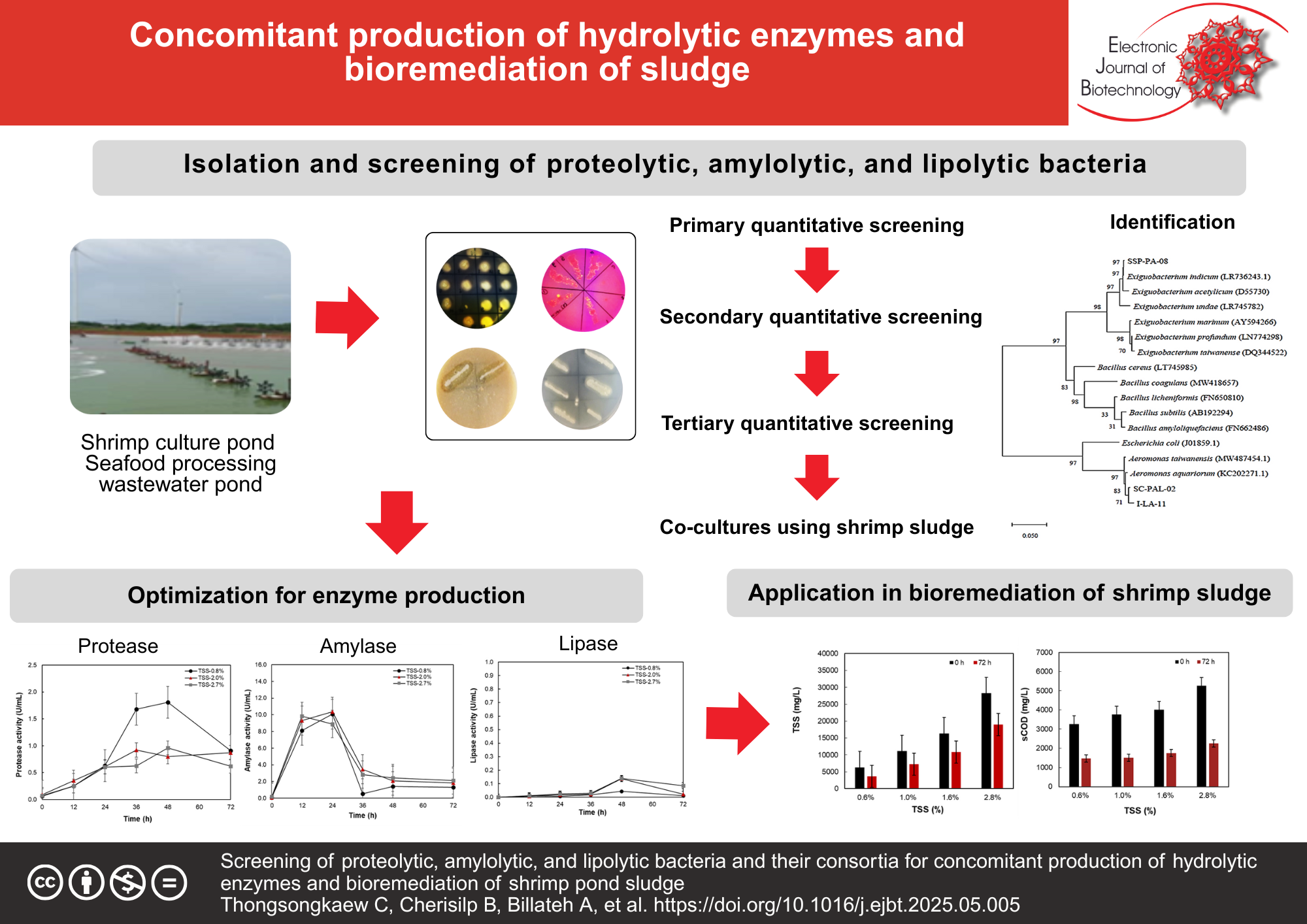
Results: The strategy using multiple substrates in the isolation media successfully obtained bacterial strains with multiple hydrolytic activities. After primary and secondary quantitative screening, 18 isolates that exhibited high dual and triple hydrolytic activities were selected. After tertiary quantitative screening using synthetic shrimp pond sludge and co-culture tests, Exiguobacterium indicum SSP-PA-08, Bacillus coagulans, and Bacillus subtilis were selected due to their synergy for the production of triple hydrolytic enzymes. Their consortia inoculated in shrimp pond sludge containing 0.8% total suspended solids (TSSs) showed an increase in proteolytic activity by 2.5 folds and amylolytic activity by 20 folds, which led to a greater reduction in TSS. The highest enzyme production was obtained using shrimp pond sludge containing 1.6% TSS.
Conclusions: This study has developed the methods to isolate bacteria with multienzyme-producing ability. Co-culturing these bacteria in synthetic shrimp sludge significantly enhanced hydrolytic activity and led to a greater reduction in TSS. These strategies may contribute greatly to the hydrolytic enzyme production and environmentally friendly bioremediation of aquaculture sludge.
References
Jasmin MY, Syukri F, Kamarudin MS, et al. Potential of bioremediation in treating aquaculture sludge. Aquaculture 2020;519:734905. https://doi.org/10.1016/j.aquaculture.2019.734905
Kamal SZ, Koyama M, Syukri F, et al. Effect of enzymatic pre-treatment on thermophilic composting of shrimp pond sludge to improve ammonia recover. Environ Res 2022;204:112299. https://doi.org/10.1016/j.envres.2021.112299 PMid: 34743806
Iber BT, Kasan NA, Recent advances in shrimp aquaculture wastewater management. Heliyon 2021;7(11):e08283. https://doi.org/10.1016/j.heliyon.2021.e08283 PMid: 34778576
Tom AP, Jayakumar JS, Biju M, et al. Aquaculture wastewater treatment technologies and their sustainability: A review. Energy Nexus 2021;4:100022. https://doi.org/10.1016/j.nexus.2021.100022
Ghattavi S, Homaei A. Marine enzymes: Classification and application in various industries. Int J Biol Macromolecules 2023;230:123136. https://doi.org/10.1016/j.ijbiomac.2023.123136 PMid: 36621739
Sharifian S, Homaei A, Kim SK, et al. Production of newfound alkaline phosphatases from marine organisms with potential functions and industrial applications. Process Biochem 2018;64:103-115. https://doi.org/10.1016/j.procbio.2017.10.005
Izadpanah Qeshmi F, Homaei A, Khajeh K, et al. Production of a novel marine Pseudomonas aeruginosa recombinant L-asparaginase: Insight on the structure and biochemical characterization. Mar Biotechnol 2022;24:599–613. https://doi.org/10.1007/s10126-022-10129-9 PMid: 35507234
Gómez S, Hurtado CF, Orellana J. Bioremediation of organic sludge from a marine recirculating aquaculture system using the polychaete Abarenicola pusilla (Quatrefages, 1866). Aquaculture 2019;507:377-384. https://doi.org/10.1016/j.aquaculture.2019.04.033
Shaik M, Sanker GG, Iswarya M, et al. Isolation and characterization of bioactive metabolites producing marine Streptomyces parvulus strain sankarensis-A10. J Genet Eng Biotechnol 2017;15(1):87-94. https://doi.org/10.1016/j.jgeb.2017.02.004 PMid: 30647645
Gozari M, Mortazavi MS, Bahador N, et al. Isolation and screening of antibacterial and enzyme-producing marine actinobacteria to approach probiotics against some pathogenic vibrios in shrimp Litopenaeus vannamei. Iran J Fish Sci 2016;15(2):630-644.
Zottig X, Meddeb-Mouelhi F, Beauregard M. Development of a high-throughput liquid state assay for lipase activity using natural substrates and rhodamine B. Anal Biochem 2016;496:25-29. https://doi.org/10.1016/j.ab.2015.11.020 PMid: 26706798
Xu X, Nielsen LJD, Song L, et al. Enhanced specificity of Bacillus metataxonomics using a tuf-targeted amplicon sequencing approach. ISME COMMUN 2023;3(1):126. https://doi.org/10.1038/s43705-023-00330-9 PMid: 38012258
Ruangwicha J, Cheirsilp B, Suyotha W. Green biorefinery of shrimp shell waste for ?-chitin and high-value co-products through successive fermentation by co-lactic acid bacteria and proteolytic fungus. Bioresour Technol 2024;393:130106. https://doi.org/10.1016/j.biortech.2023.130106 PMid: 38008224
Jain A, Jain R, Jain S. Quantitative analysis of reducing sugars by 3, 5-Dinitrosalicylica acid (DNSA Method). In: Basic Techniques in Biochemistry, Microbiology and Molecular Biology. Springer Protocols Handbooks. Humana, New York, NY 2020. https://doi.org/10.1007/978-1-4939-9861-6_43
Louhasakul Y, Cheirsilp B. Potential use of industrial by-products as promising feedstock for microbial lipid and lipase production and direct transesterification of wet yeast into biodiesel by lipase and acid catalysts. Bioresour Technol 2022;348:126742. https://doi.org/10.1016/j.biortech.2022.126742 PMid: 35065222
Ibrahim ASS, Al-Salamah AA, Elbadawi YB, et al. Production of extracellular alkaline protease by new halotolerant alkaliphilic Bacillus sp. NPST-AK15 isolated from hyper saline soda lakes. Electron J Biotechnol 2015;18(3):236-243. https://doi.org/10.1016/j.ejbt.2015.04.001
Kot AM, B?a?ejak S, Kurcz A, et al. Biodegradation of deproteinized potato wastewater and glycerol during cultivation of Rhodotorula glutinis yeast. Electro J Biotechnol 2015;18(6):428-432. https://doi.org/10.1016/j.ejbt.2015.08.006
Panigrahi A, Saranya C, Sundaram M, et al. Carbon, nitrogen (C/N) ratio level variation influences microbial community of the system and growth as well as immunity of shrimp (Litopenaeus vannamei) in biofloc based culture system. Fish Shellfish Immunol 2018;81:329-337. https://doi.org/10.1016/j.fsi.2018.07.035 PMid: 30016684
Al-Dhabi NA, Esmail GA, Mohammed Ghilan AK, et al. Isolation and screening of Streptomyces sp. Al-Dhabi-49 from the environment of Saudi Arabia with concomitant production of lipase and protease in submerged fermentation. Saudi J Biol Sci 2020;27(1):474-479. https://doi.org/10.1016/j.sjbs.2019.11.011 PMid: 31889873
Gaona CAP, de Almeida MS, Viau V, et al. Effect of different total suspended solids levels on a Litopenaeus vannamei (Boone, 1931) BFT culture system during biofloc formation. Aquac Res 2017;48(3):1070-1079. https://doi.org/10.1111/are.12949
Solangi F, Zhu X, Solangi KA, et al. Responses of soil enzymatic activities and microbial biomass phosphorus to improve nutrient accumulation abilities in leguminous species. Sci Rep 2024;14:11139. https://doi.org/10.1038/s41598-024-61446-z PMid: 38750151
Soaudy MR, Ghonimy A, Greco LSL, et al. Total suspended solids and their impact in a biofloc system: Current and potentially new management strategies. Aquacult 2023;572:739524. https://doi.org/10.1016/j.aquaculture.2023.739524
Kumar SS, Jithin V, Jijeesh V, et al. Production and purification of alkaline protease from Exiguobacterium indicum TBG-PICH-001 isolated from soil samples of Pichavaram Estuary (Tamil Nadu). Indian J Geo-Mar Sci 2018;47(3):580-586.
Yasin MT, Ali Y, Ahmad K, et al. Alkaline lipase production by novel meso-tolerant psychrophilic Exiguobacterium sp. strain (AMBL-20) isolated from glacier of northeastern Pakistan. Arch Microbiol 2021;203(4):1309-1320. https://doi.org/10.1007/s00203-020-02133-1 PMid: 33325000
Ramamurthy V, Cheepurupalli L, Singh SR, et al. Co-culture:A promising method in enzyme production. Int J Chemtech Res 2017;10(6):720-726.
Saelor A, Kongjan P, Prasertsan P, et al. Enhancing the efficiency of high solid anaerobic digestion of empty fruit bunches under thermophilic conditions by particle size reduction and co-digestion with palm oil mill effluent. Carbon Resour Conver 2024;8(2):100262. https://doi.org/10.1016/j.crcon.2024.100262
Ramu D, Sigamani S, Venkatachalam H, et al. The role of probiotics in the control of bacterial diseases and biodegradation of organic matter in shrimp (Penaeus vannamei) culture ponds of South India. J Coastal Life Med 2017;5(7):293-298. https://doi.org/10.12980/jclm.5.2017J7-32
Cong M, Jiang Q, Xu X, et al. The complete genome sequence of Exiguobacterium arabatum W?01 reveals potential probiotic functions. MicrobiologyOpen 2017;6(5):e00496. https://doi.org/10.1002/mbo3.496 PMid: 28589562
Hakim A, Bhuiyan FR, Iqbal A, et al. Production and partial characterization of dehairing alkaline protease from Bacillus subtilis AKAL7 and Exiguobacterium indicum AKAL11 by using organic municipal solid wastes. Heliyon 2018;4(6):e00646. https://doi.org/10.1016/j.heliyon.2018.e00646 PMid: 30009270
Bahaddad SA, Almalki MHK, Alghamdi OA, et al. Bacillus species as direct-fed microbial antibiotic alternatives for monogastric production. Probiotics Antimicrob Proteins 2023;15(1):1-16. https://doi.org/10.1007/s12602-022-09909-5 PMid: 35092567
Lázaro-Mass S, Gómez-Cornelio S, Castillo-Vidal M, et al. Biodegradation of hydrocarbons from contaminated soils by microbial consortia: A laboratory microcosm study. Electron J Biotechnol 2023;61:24-32. https://doi.org/10.1016/j.ejbt.2022.10.002
Cao Z, Yan W, Ding M, et al. Construction of microbial consortia for microbial degradation of complex compounds. Front Bioeng Biotechnol 2022;10:1051233. https://doi.org/10.3389/fbioe.2022.1051233 PMid: 36561050
Safitri R, Priadie B, Miranti M, Astuti AW, Ability of bacterial consortium: Bacillus coagulans, Bacillus licheniformis, Bacillus pumilus, Bacillus subtilis, Nitrosomonas sp., and Pseudomonas putida in bioremediation of wastewater in Cisirung wastewater treatment plant. AgroLife Sci J 2015;4(1):146-152.
Kandasamy S, Muthusamy G, Balakrishnan S, et al. Optimization of protease production from surface-modified coffee pulp waste and corncobs using Bacillus sp. by SSF. 3 Biotech 2016;6:167. https://doi.org/10.1007/s13205-016-0481-z PMid: 28330239
Sial A, Zhang B, Zhang A, et al. Microalgal–bacterial synergistic interactions and their potential influence in wastewater treatment: A review. Bioenergy Res 2021;14(3):723-738. https://doi.org/10.1007/s12155-020-10213-9
Nzila A, Razzak SA, Zhu J. Bioaugmentation, an emerging strategy of industrial wastewater treatment for reuse and discharge. Int J Environ Res Public Health 2016;13(9):846. https://doi.org/10.3390/ijerph13090846 PMid: 27571089
Madariaga ST, Marín SL. Sanitary and environmental conditions of aquaculture sludge. Aquacule Res 2017;48(4): 1744-1750. https://doi.org/10.1111/are.13011

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Copyright (c) 2025 Electronic Journal of Biotechnology